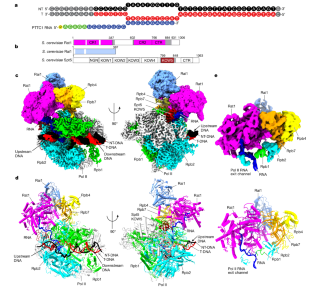
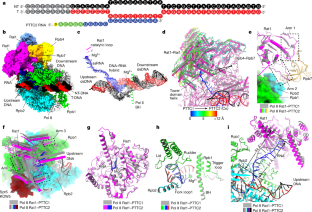
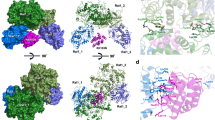
Efficient termination is required for robust gene transcription. Eukaryotic organisms use a conserved exoribonuclease-mediated mechanism to terminate the mRNA transcription by RNA polymerase II (Pol II) 1,2,3,4,5 . Here we report two cryogenic electron microscopy structures of Saccharomyces cerevisiae Pol II pre-termination transcription complexes bound to the 5′-to-3′ exoribonuclease Rat1 and its partner Rai1. Our structures show that Rat1 displaces the elongation factor Spt5 to dock at the Pol II stalk domain. Rat1 shields the RNA exit channel of Pol II, guides the nascent RNA towards its active centre and stacks three nucleotides at the 5′ terminus of the nascent RNA. The structures further show that Rat1 rotates towards Pol II as it shortens RNA. Our results provide the structural mechanism for the Rat1-mediated termination of mRNA transcription by Pol II in yeast and the exoribonuclease-mediated termination of mRNA transcription in other eukaryotes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
196,21 € per year
only 3,85 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex
Article Open access 08 September 2024
Structures illustrate step-by-step mitochondrial transcription initiation
Article Open access 11 October 2023

Structural basis of Integrator-dependent RNA polymerase II termination
Article Open access 03 April 2024
Data availability
Atomic coordinates have been deposited in the PDB under the accession codes 8JCH (Pol II Rat1–PTTC1) and 8K5P (Pol II Rat1–PTTC2). Corresponding cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under the accession codes EMD-36162 (Pol II Rat1–PTTC1) and EMD-36908 (Pol II Rat1–PTTC2). Source data are provided with this paper.
References
- Kim, M. et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature432, 517–522 (2004). ArticleCASPubMedADSGoogle Scholar
- West, S., Gromak, N. & Proudfoot, N. J. Human 5′ -> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature432, 522–525 (2004). ArticleCASPubMedADSGoogle Scholar
- Luo, W. & Bentley, D. A ribonucleolytic rat torpedoes RNA polymerase II. Cell119, 911–914 (2004). ArticleCASPubMedGoogle Scholar
- Fong, N. et al. Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol. Cell60, 256–267 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Krzyszton, M. et al. Defective XRN3-mediated transcription termination in Arabidopsis affects the expression of protein-coding genes. Plant J.93, 1017–1031 (2018). ArticleCASPubMedGoogle Scholar
- Proudfoot, N. J. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science352, aad9926 (2016). ArticlePubMedPubMed CentralGoogle Scholar
- Girbig, M., Misiaszek, A. D. & Muller, C. W. Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases. Nat. Rev. Mol. Cell Biol.23, 603–622 (2022). ArticleCASPubMedGoogle Scholar
- Porrua, O. & Libri, D. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat. Rev. Mol. Cell Biol.16, 190–202 (2015). ArticleCASPubMedGoogle Scholar
- Eaton, J. D. & West, S. Termination of transcription by RNA polymerase II: BOOM! Trends Genet.36, 664–675 (2020). ArticleCASPubMedGoogle Scholar
- Richard, P. & Manley, J. L. Transcription termination by nuclear RNA polymerases. Genes Dev.23, 1247–1269 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kumar, A., Clerici, M., Muckenfuss, L. M., Passmore, L. A. & Jinek, M. Mechanistic insights into mRNA 3′-end processing. Curr. Opin. Struc. Biol.59, 143–150 (2019). ArticleCASGoogle Scholar
- Lunde, B. M. et al. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol.17, 1195–1201 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Carminati, M. et al. A direct interaction between CPF and RNA Pol II links RNA 3′ end processing to transcription. Mol. Cell83, 4461–4478 (2023).
- Brannan, K. et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol. Cell46, 311–324 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Nojima, T. et al. Mammalian NET-Seq reveals genome-wide nascent transcription coupled to RNA processing. Cell161, 526–540 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Jimeno-Gonzalez, S., Haaning, L. L., Malagon, F. & Jensen, T. H. The yeast 5′-3′ exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Mol. Cell37, 580–587 (2010). ArticleCASPubMedGoogle Scholar
- Cortazar, M. A. et al. Xrn2 substrate mapping identifies torpedo loading sites and extensive premature termination of RNA pol II transcription. Genes Dev.36, 1062–1078 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kim, M., Ahn, S. H., Krogan, N. J., Greenblatt, J. F. & Buratowski, S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J.23, 354–364 (2004). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhang, Z., Fu, J. & Gilmour, D. S. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev.19, 1572–1580 (2005). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhang, H., Rigo, F. & Martinson, H. G. Poly(A) signal-dependent transcription termination occurs through a conformational change mechanism that does not require cleavage at the poly(A) site. Mol. Cell59, 437–448 (2015). ArticleCASPubMedGoogle Scholar
- Logan, J., Falck-Pedersen, E., Darnell, J. E. Jr & Shenk, T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc. Natl Acad. Sci. USA84, 8306–8310 (1987). ArticleCASPubMedPubMed CentralADSGoogle Scholar
- Eaton, J. D., Francis, L., Davidson, L. & West, S. A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes. Genes Dev.34, 132–145 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Luo, W., Johnson, A. W. & Bentley, D. L. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric–torpedo model. Genes Dev.20, 954–965 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Cortazar, M. A. et al. Control of RNA Pol II speed by PNUTS-PP1 and Spt5 dephosphorylation facilitates termination by a “sitting duck torpedo” mechanism. Mol. Cell76, 896–908 e894 (2019). ArticleCASPubMedPubMed CentralGoogle Scholar
- Xue, Y. et al. Saccharomyces cerevisiaeRAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol.20, 4006–4015 (2000). ArticleCASPubMedPubMed CentralGoogle Scholar
- Xiang, S. et al. Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature458, 784–788 (2009). ArticleCASPubMedPubMed CentralADSGoogle Scholar
- Mayer, A. et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol.17, 1272–1278 (2010). ArticleCASPubMedGoogle Scholar
- Narain, A. et al. Targeted protein degradation reveals a direct role of SPT6 in RNAPII elongation and termination. Mol. Cell81, 3110–3127 (2021). ArticleCASPubMedPubMed CentralGoogle Scholar
- Baejen, C. et al. Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol. Cell66, 38–49 (2017). ArticleCASPubMedGoogle Scholar
- Jinek, M., Coyle, S. M. & Doudna, J. A. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell41, 600–608 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Stevens, A. & Poole, T. L. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J. Biol. Chem.270, 16063–16069 (1995). ArticleCASPubMedGoogle Scholar
- Ehara, H. et al. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science357, 921–924 (2017). ArticleCASPubMedADSGoogle Scholar
- Parua, P. K. et al. A Cdk9–PP1 switch regulates the elongation–termination transition of RNA polymerase II. Nature558, 460–464 (2018). ArticleCASPubMedPubMed CentralADSGoogle Scholar
- Kecman, T. et al. Elongation/termination factor exchange mediated by PP1 phosphatase orchestrates transcription termination. Cell Rep.25, 259–269 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Grosso, A. R., de Almeida, S. F., Braga, J. & Carmo-Fonseca, M. Dynamic transitions in RNA polymerase II density profiles during transcription termination. Genome Res.22, 1447–1456 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Orozco, I. J., Kim, S. J. & Martinson, H. G. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J. Biol. Chem.277, 42899–42911 (2002). ArticleCASPubMedGoogle Scholar
- Gromak, N., West, S. & Proudfoot, N. J. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol.26, 3986–3996 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- James, K., Gamba, P., Cockell, S. J. & Zenkin, N. Misincorporation by RNA polymerase is a major source of transcription pausing in vivo. Nucleic Acids Res.45, 1105–1113 (2017). CASPubMedGoogle Scholar
- Molodtsov, V., Wang, C., Firlar, E., Kaelber, J. T. & Ebright, R. H. Structural basis of Rho-dependent transcription termination. Nature14, 367–374 (2023). ArticleADSGoogle Scholar
- Murayama, Y. et al. Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus. Sci. Adv.9, eade7093 (2023). ArticleCASPubMedPubMed CentralGoogle Scholar
- Chengyuan, W. et al. Structural basis of archaeal FttA-dependent transcription termination. Preprint at bioRxivhttps://doi.org/10.1101/2023.08.09.552649 (2023).
- Fianu, I. et al. Structural basis of integrator-mediated transcription regulation. Science374, 883–887 (2021). ArticleCASPubMedADSGoogle Scholar
- Zheng, H. et al. Structural basis of INTAC-regulated transcription. Protein Cell14, 698–702 (2023). ArticlePubMedPubMed CentralGoogle Scholar
- Higo, T. et al. Development of a hexahistidine-3x FLAG-tandem affinity purification method for endogenous protein complexes in Pichia pastoris. J. Struct. Funct. Genomics15, 191–199 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods14, 331–332 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol.193, 1–12 (2016). ArticleCASPubMedPubMed CentralADSGoogle Scholar
- Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife7, e42166 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). ArticleCASPubMedGoogle Scholar
- Xu, J. et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature551, 653–657 (2017). ArticleCASPubMedPubMed CentralADSGoogle Scholar
- Farnung, L., Ochmann, M., Engeholm, M. & Cramer, P. Structural basis of nucleosome transcription mediated by Chd1 and FACT. Nat. Struct. Mol. Biol.28, 382–387 (2021). ArticleCASPubMedPubMed CentralGoogle Scholar
- Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). ArticleCASPubMedPubMed CentralADSGoogle Scholar
- Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem.25, 1605–1612 (2004). ArticleCASPubMedGoogle Scholar
- Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D60, 2126–2132 (2004). ArticlePubMedADSGoogle Scholar
- Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221 (2010). ArticleCASPubMedPubMed CentralADSGoogle Scholar
- Ehara, H., Kujirai, T., Shirouzu, M., Kurumizaka, H. & Sekine, S. I. Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT. Science377, eabp9466 (2022). ArticleCASPubMedGoogle Scholar
Acknowledgements
The work was supported by the National Key Research and Development Program of China 2018YFA0900701 (Y. Zhang) and the Basic Research Zone Program of Shanghai JCYJ-SHFY-2022-012 (Y. Zhang). We thank L. Kong, F. Wang, G. Li, J. Duan at electron microscopy system at the National Facility for Protein Science in Shanghai (NFPS), M. Zhang at the electron microscopy center in Interdisciplinary Research Center on Biology and Chemistry (IRCBC), and staffs X. Men, F. Liu, and S. Wang at electron microscopy center at Shuimu Biosciences in Hangzhou for providing technical support and assistance in cryo-EM data collection. We thank M. Zhang and Z. Zhang at the core facility of the Center for Excellence in Molecular Plant Sciences (CEMPS) for technical support and assistance in screening cryo-EM samples. The S. cerevisiae BJ2168 strain was a gift from G. Cai. The pESC-his vector was a gift from Z. Zhou.
Author information
Authors and Affiliations
- Key Laboratory of Synthetic Biology, National Key Laboratory of Plant Design, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China Yuan Zeng, Hong-Wei Zhang, Xiao-Xian Wu & Yu Zhang
- University of Chinese Academy of Sciences, Beijing, China Yuan Zeng & Hong-Wei Zhang
- Yuan Zeng









